Usage¶
Installation¶
This software can be used in a few different ways. To run processing workflows, called ‘procedures’, there is a command line cli, that resides in main.py and runs workflow files on given CTD data files.
The easiest way to install CLI interface called ‘ctdpro’ is to use pipx:
$ pipx install ctd-processing
$ ctdpro check
All set, you are ready to go.
Otherwise, via basic python package managers, like pip, conda or poetry:
# using pip
$ python -m venv .venv
$ source .venv/bin/activate
$ pip install ctd-processing
$ python src/processing/main.py check
All set, you are ready to go.
# using poetry
$ poetry install
$ poetry env activate
$ ctdpro check
All set, you are ready to go.
CLI¶
The CLI ‘ctdpro’ should be the main entry point to the software. It allows running and editing of procedure workflows. It features the following commands:
$ ctpro --help
Usage: ctdpro [OPTIONS] COMMAND [ARGS]...
╭─ Options ────────────────────────────────────────────────────────────────────────────────────────────────────────╮
│ --install-completion Install completion for the current shell. │
│ --show-completion Show completion for the current shell, to copy it or customize the installation. │
│ --help Show this message and exit. │
╰──────────────────────────────────────────────────────────────────────────────────────────────────────────────────╯
╭─ Commands ───────────────────────────────────────────────────────────────────────────────────────────────────────╮
│ run Processes one target file using the given procedure workflow file. │
│ convert Converts a list of Sea-Bird raw data files (.hex) to .cnv files. │
│ batch Applies a processing config to multiple .hex or. cnv files. │
│ edit Opens a procedure workflow file in GUI for editing. │
│ show Display the contents of a procedure workflow file. │
│ check Assures that all requirements to use this tool are met. │
╰──────────────────────────────────────────────────────────────────────────────────────────────────────────────────╯
The top three commands are the commands to run processing workflows, while the other ones are for editing workflow files and general checking whether the requirements are met.
To get more specific information to the individual commands and their arguments, you can display help sites for each of them:
$ ctdpro batch --help
Usage: ctdpro batch [OPTIONS] INPUT_DIR CONFIG
Applies a processing config to multiple .hex or. cnv files.
Parameters ---------- input_dir: Path | str : The data directory with the target files. config: dict | Path | str: Either an explicit config as dict or a path to a .toml config file. pattern: str : A
name pattern to filter the target files with. (Default is ".cnv")
Returns ------- A list of paths or CnvFiles of the processed files.
╭─ Arguments ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────╮
│ * input_dir TEXT [default: None] [required] │
│ * config TEXT [default: None] [required] │
╰───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────╯
╭─ Options ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────╮
│ --pattern TEXT [default: .cnv] │
│ --help Show this message and exit. │
╰───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────╯
Intro¶
The workflow files configure everything needed to run a processing procedure and are easy to understand and write.
Another way is to run processing procedures directly from your python code, using either workflow files or the fact that these workflows correspond to a basic python dictionary internally. This allows for quick scripts that can use flexible input variables.
Warning
To run Sea-Bird processing modules, you need a local installation of them, from their website.
from processing import Procedure
# using own dictionary:
proc_config = {
"psa_directory": "path_to_psas",
"output_dir": "path_to_output",
"modules": {
"datcnv": {"psa": "DatCnv.psa"},
"wildedit_geomar": {"std1": 3.0, "window_size": 100},
"celltm": {"psa": ("CellTM.psa")},
},
}
procedure = Procedure(proc_config)
# using a .toml workflow file:
procedure = Procedure("example_config.toml")
Workflow files¶
The workflow dictionary used in the example above corresponds to this .toml workflow file:
# example_config.toml
psa_directory = "path_to_psas"
output_dir = "path_to_output"
[modules.datcnv]
psa = "DatCnv.psa"
[modules.wildedit_geomar]
std1 = 3.0
window_size = 100
[modules.celltm]
psa = "CellTM.psa"
It does not matter, which way you provide the processing workflow information, the two above examples work in exactly the same way. The different options available and how to use them, are shown below:
option |
type in .toml |
type in dictionary |
description |
|---|---|---|---|
input |
str |
Path, str, CnvFile |
CnvFile instance or path to a file |
psa_directory |
str |
Path, str |
Path to a directory |
output_dir |
str |
Path, str |
Path to a directory |
output_name |
str |
str |
The new name of the output file |
output_type |
str |
str |
“cnv” for a .cnv file or anything else for a CnvFile instance |
xmlcon |
str |
Path, str |
Path to a .xmlcon file |
modules |
dict |
dict |
A dictionary containing the processing modules and their options |
Tip
The only mandatory option is the modules dictionary, describing the steps to run on the file(s). All other options are optional.
A simple gui can be used to write and edit workflow files, which, at the moment, is run like this:
ctdpro edit path_to_toml.toml
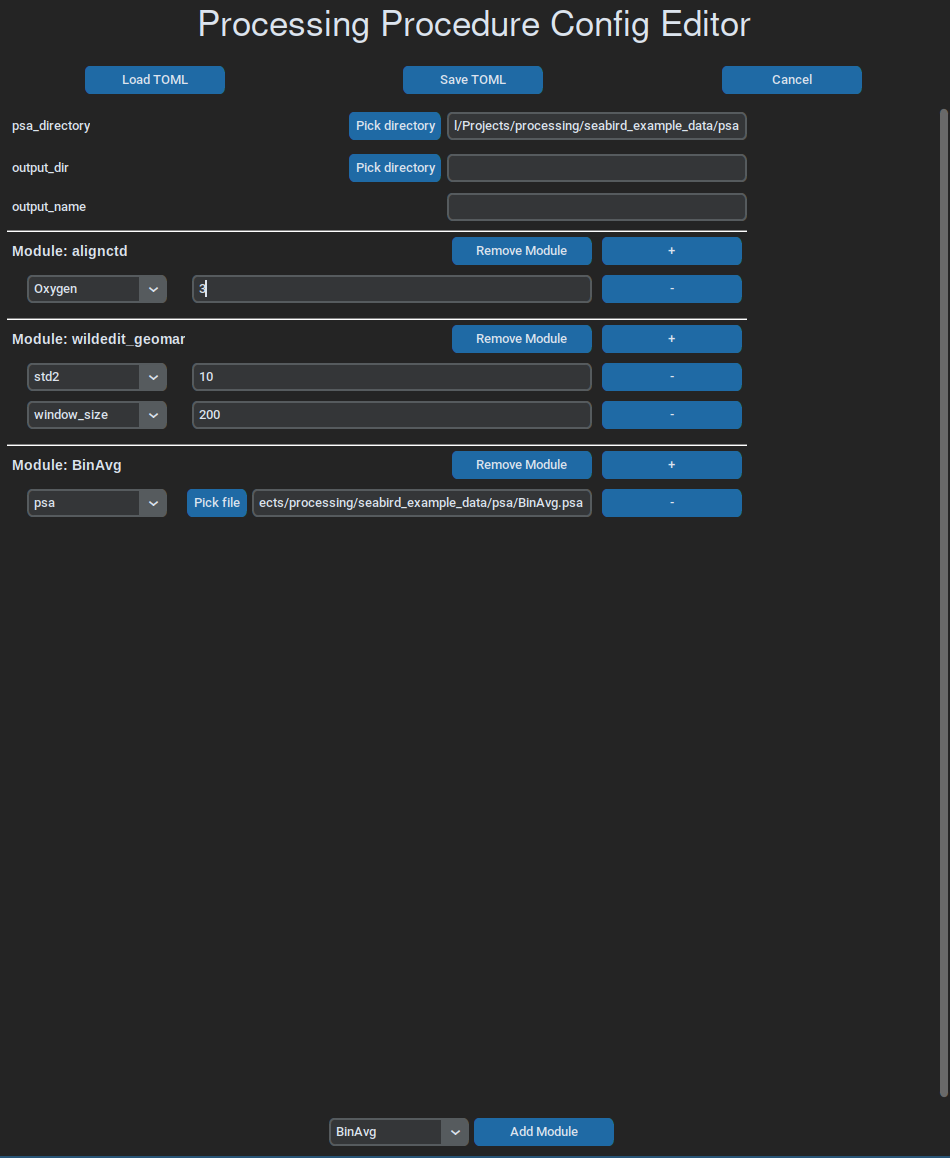
Batch Processing¶
There are also functions for batch processing, that allow for quick processing of multiple target files, using the very same workflow configuration.
from processing import convert, batch_processing
# converting multiple files
list_of_converted_files = convert(
input_dir="path_to_hex_files",
psa_path="path_to_psas",
)
# general batch processing on multiple files, using the same config
list_of_processed_files = batch_processing(
input_dir="path_to_input_files",
config="path_to_toml_config",
)
You can of course just do flexible batch processing inside a loop, like shown in following example, running inside a jupyter notebook. This is a common way of doing interactive ctd data processing, using this package.
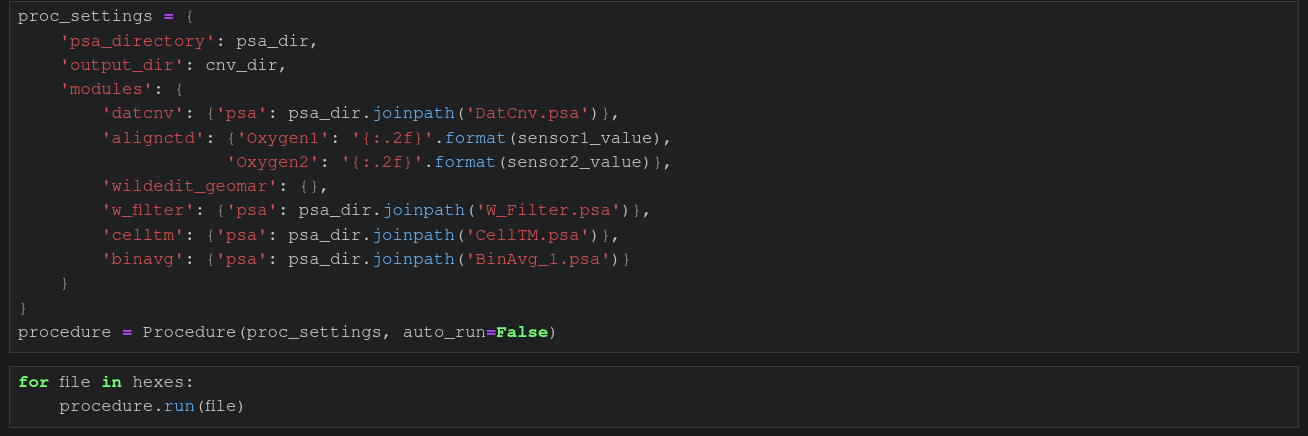
Note
All options for the Procedure class can be found here.
Outlook¶
One development goal remains to rewrite all Sea-Bird processing modules as more modern, flexible python modules, which would lead to an end of all the file reading and writing processes, as all the data processing steps could run on one internal python object, representing a Sea-Bird data file. We will stick to this file architecture to ensure future compatibility with other processing workflows, relying on the original Sea-Bird processing modules. Another great result of this rewrite would be the end of the .psa configuration files, which are bloated and a pain to maintain between changes of processing workflows.
Additionally, new modules are going to be written regularly, with the currently available ones found here.